
How to query KVarPredDB
In order to facilitate data interpretation, graphical representations with interactive features were used as required. Querying the database is primarily based on either “cDNA Variant” or “Disease” under the “Search” menu.
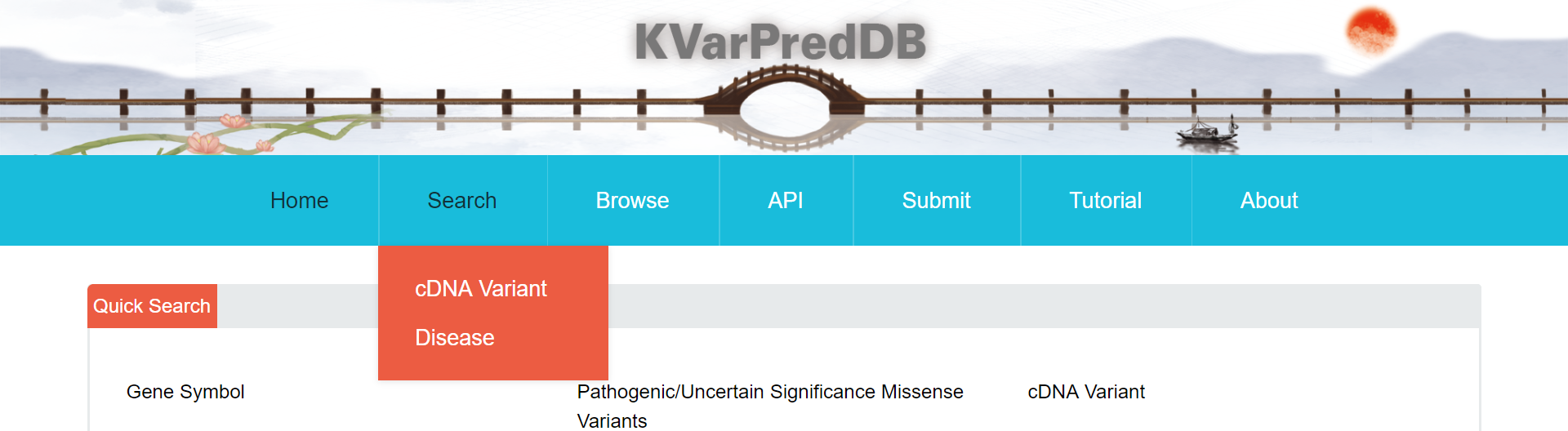
From “cDNA Variant” page, users can quickly access the database by choosing “Gene Symbol”, “Uncertain Significance Missense Variants” or “Pathogenic Missense Variants” which are implemented by a drop-down list. Here, the term ‘Uncertain Significance Missense Variants’ represents the Pathogenicity of DNA variation in populations is unknown while the term 'Pathogenic Missense Variants' indicates cases there is a reasonable suspicion that the variant is deleterious. For each data of Pathogenic Missense Variants, lollipop plot, related diseases, molecular docking results, related papers are displayed by default. Lollipop plot is used to Map mutations on a linear protein and its domains; For the related disease, a link to OMIM database is provided; Box tab in Molecular Docking Results section visualizes the binding energy difference between wild and mutation, and the data is available for the download. The changes is visualized by a movie generated by Pymol. An embedded module from RCSC supported by NGL viewer was employed to display the molecular graphics.
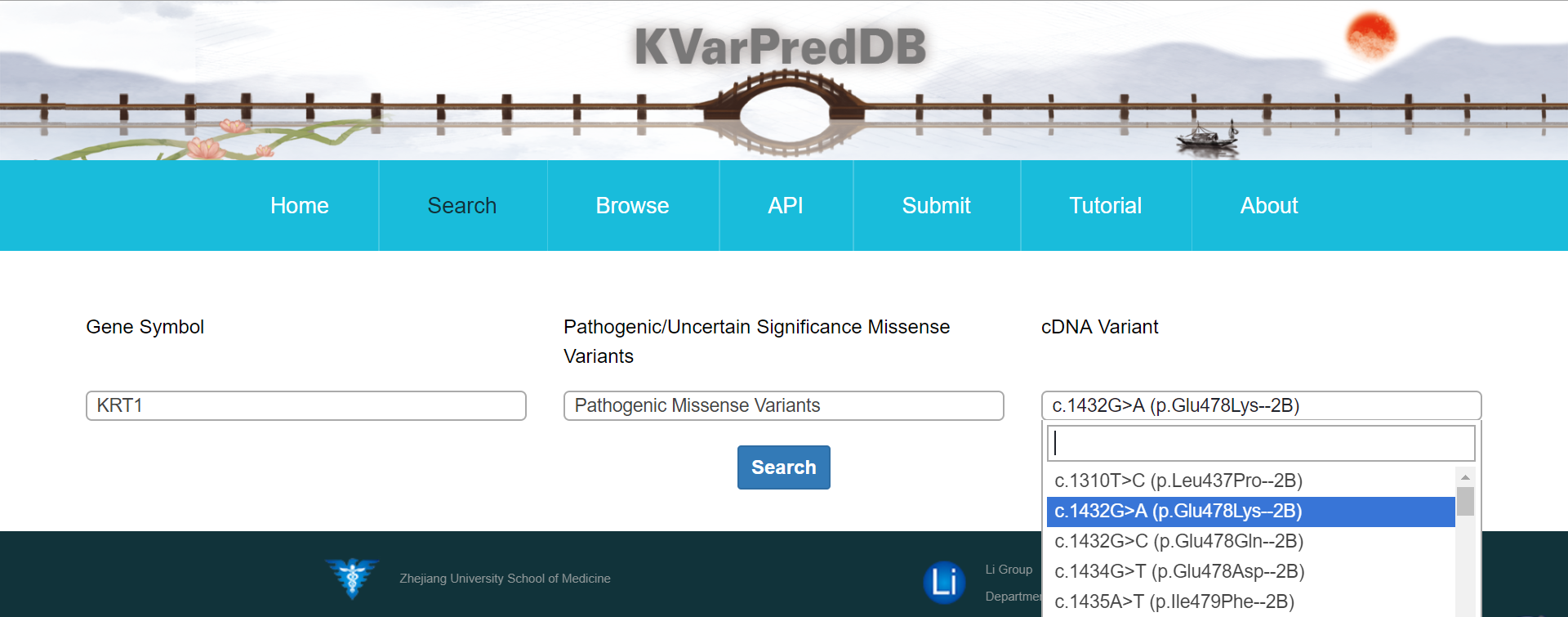
Likewise, browsing the “Disease” page leads directly to disease description.
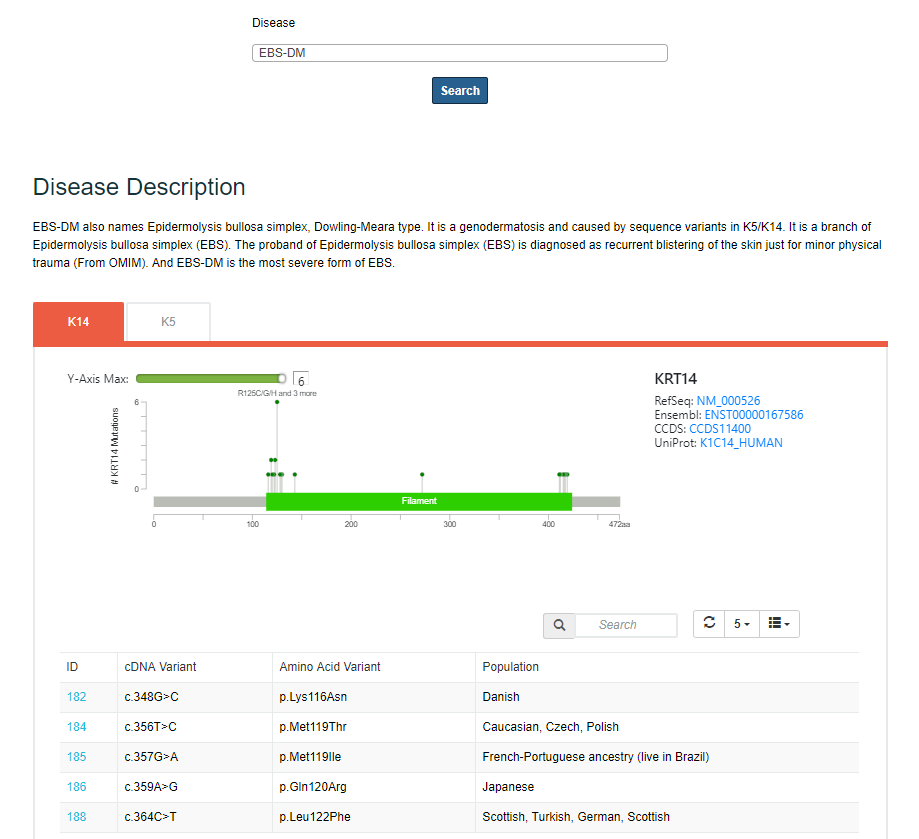
After you selecting the disease type you are interested, the page will show a lollipop plot summarizing the mutations on the domains of individual keratin gene. Details of these mutations are given in a tabular collection format.
Under “Browse” menu, the user can navigate the pages of “Keratin Genes/Proteins”, “Disease List” and “Protein structure & Molecular Docking” which summarize all related details and each entry can be linked to individual pages.
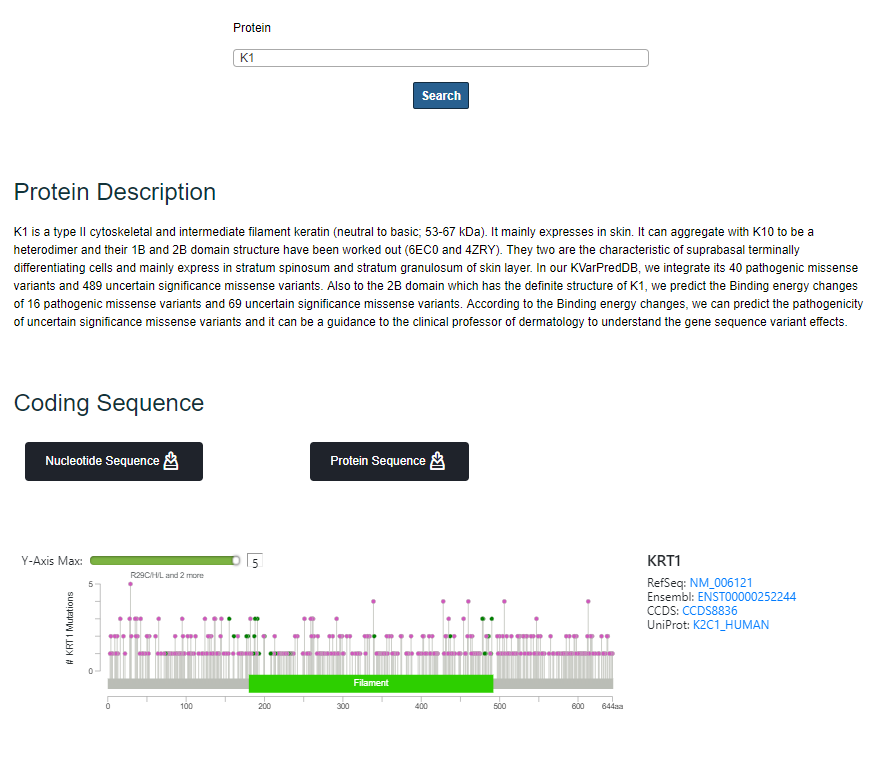
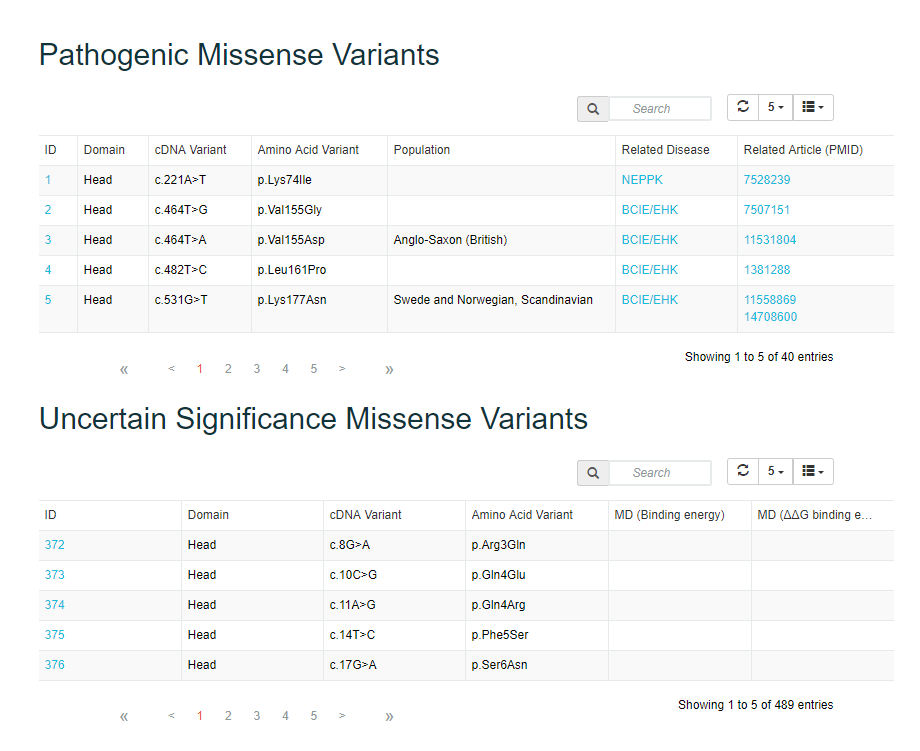
After selecting the protein when the user chooses “Keratin Genes/Proteins”, a brief introduction of the protein, lollipop plot summarizing the mutations on the domains, tabular list of Pathogenic Missense Variantss (including information about Domain, cDNA variant, AminoAcid Variant, Population, Related Disease, Related Article) and Uncertain Significance Missense Variants (Domain, cDNA Variant, Amino Acid Variant, MD Binding energy and MD ddG binding energy) are shown as default.
Browsing the “Disease List” displays the 25 diseases in our database. The molecular Docking page showed the 3TNU and 4ZRY crystal structure and the related variant on them. For each variant, ID, protein, cDNA Variant, Mutation, Binding energy and ΔΔG binding energy are displayed by default.
Browsing the “Disease List” , the page will display all the 25 diseases in our database, which could be hyperlinked to OMIM database. The “Protein Structure & Molecular Docking” page showed the crystal structures of 3TNU, 4ZRY and 6EC0 crystal structure and the related information of the variants. For each variant, ID, protein, cDNA Variant, Mutation, Binding energy and ΔΔG binding energy are displayed by default.